Remap: A reprojection tool of geolocated data between various grids
Language/Format: C, C++
Application type(s): Reprojection
Related project(s): CALIPSO, MODIS, MSG
NAME
remap
DOWNLOAD
SOURCE FILES
SYNOPSIS
remap [-option …] dataset_ref@file_ref dataset_to_remap_1@file_1 dataset_to_remap_2@file_2 …
DESCRIPTION
remap ensures reprojection of geolocated data between various grids of satellite-embedded instruments.
The first argument is the user’s chosen reference dataset. A dataset is specified by its name followed by the name of the file where it is to be found, using the syntax : dataset@file. The dataset is expected to be a 2-dimensional array. Following dataset arguments will be remapped over the reference dataset geometry (using a simple nearest-pixel policy for the time being, more clever reprojection policies may be implemented in future releases). Reprojected datasets will be put in a common HDF file, along with the reference dataset and its geolocation data (latitudes, longitudes and acquisition times of each reference pixel). Distances and time differences between reprojected pixels and their references may also be stored at the user’s will.
The user is given the ability to rename datasets to give them better names in the output file. This functionality may also be used to resolve collisions between input dataset names. The syntax is straightforward: old_dataset_name/new_dataset_name@input_file
In a few cases (e.g., MODIS), some datasets are 3-dimensional arrays, which are stacks of 2-dimensional arrays indexed by a plane number (typically a channel number). In this case, the user is to specify the plane number between brackets, e.g. dataset[1]@file. The convention adopted here is that the first plane is indexed by one, not by zero.
Higher-dimensional datasets are currently not supported.
For the time being, only a few satellite products are supported, namely MODIS (on AQUA and TERRA), IIR (on CALIPSO) and SEVIRI (on METEOSAT-8). More products are intended to be integrated in the future (an issue is the diversity of formats and conventions in this applicative domain)
remap will yeld an exit code of zero on success. Any other value means failure. Error messages are to be sent to the standard error output. Verbose messages (displayed when the option -v is set) will be sent to the standard output.
EXAMPLE
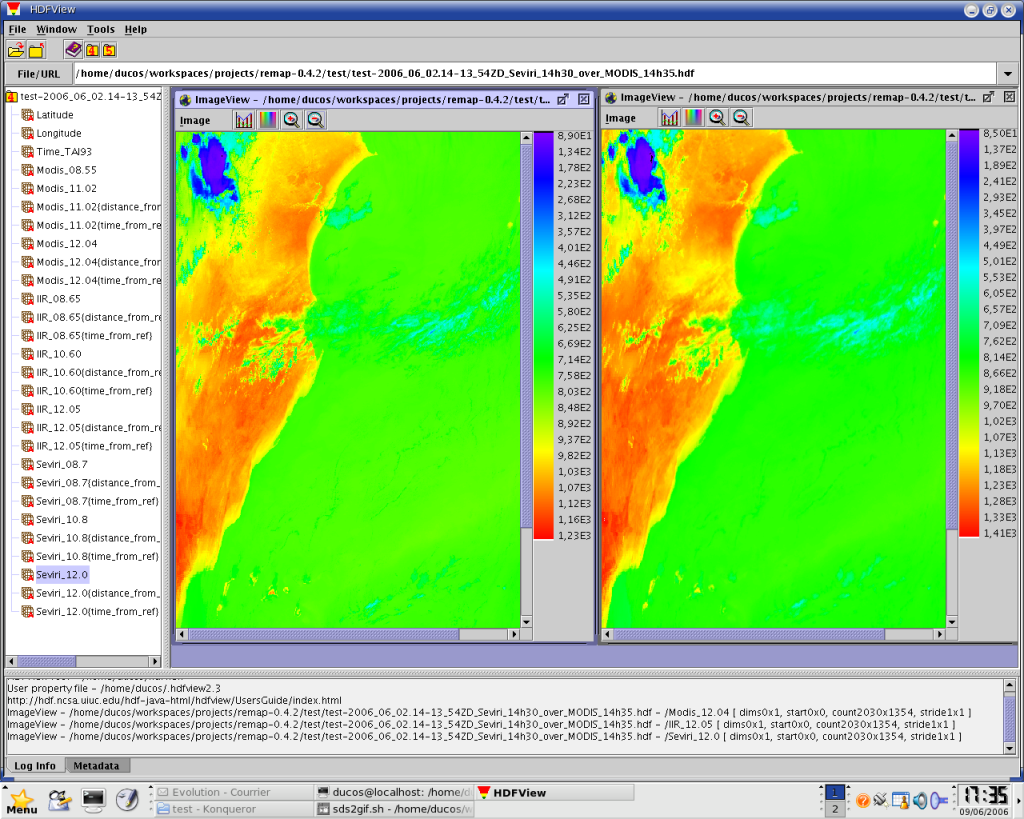
an output of remap, viewed with HDFView : the SEVIRI 12.0 micrometers channel (on the left) has been remapped over the MODIS 12.04 micrometers channel (on the right)
OPTIONS
Options of remap :
- -c slope,offset
- calibration factors (default: slope = 0.01, offset = 0)
- -d distance
- distance of colocalization, in km (default is 1 km)
- -h
- advanced help (displays supported types and implemented user defined projection)
- -m <specification for missing value>
- specifies values for missing coordinates, time or data (use -h for details)
- -p user_defined_grid
- specifies a user defined projection instead of a file defined projection (use -h for details)
- -o filename
- output filename (default is remap.out.hdf)
- -t max_dtime
- max difference in time from the pixels of reference, in minutes (may be inf, for infinite, or a strictly positive real number, default is inf)
- -v
- verbose mode
- -x
- extended output (with differences in time and distances from references recorded)
FILES
Currently supported input products include:
- MODIS/TERRA
- MOD021??.A???????.????.???.?????????????*.hdf
- MODIS/AQUA
- MYD021??.A???????.????.???.?????????????*.hdf
- IIR/CALIPSO
- CAL_IIR_L1-*.hdf
- SEVIRI
- H-000-MSG1__-MSG1________-?????????-CYCLE____-????????????
(each ? means exactly one character, and * any sequence of characters, maybe empty)
ENVIRONMENT
REMAP_RAWPATH path to a directory containing SEVIRI geolocation files, namely seviri_latitudes.raw and seviri_longitudes.raw (indeed, SEVIRI geolocation data are not available in products as they never change).
COPYRIGHT
This program is free software; you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation; either version 2 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program; if not, write to the Free Software Foundation, Inc., 51 Franklin St, Fifth Floor, Boston, MA 02110-1301 USA
BUGS
This is an early version of the software. While main functionalities are implemented and the software may be used as is to perform remapping, a lot of bugs surely still wait to get revealed.
Please report any bug you could find to fabrice.ducos@univ-lille.fr